About the TriCleanse™ Process
The Power of Decellularization
The proper decellularization of ECMs is critical to the function of the final ECM product. The goal of decellularization is to remove the cells and eliminate residual genetic material, while preserving the ECM components and retaining the properties of the base tissue.1 Incomplete decellularization has been shown to decrease the ratio of M2-activated macrophages to M1-macrophages when compared to more complete decellularization methods.2 Further studies have demonstrated that higher ratios of M2-activated to M1-activated macrophages are more promotive of immunomodulation, constructive mechanisms, and remodeling activities.3
Decellularization Efficiency of the TriCleanse™ Process
Placental devices manufactured utilizing the TriCleanse™ Process were compared to raw untreated placentas to determine the decellularization efficiency of the process. Histologic evaluations were performed to assess the impact of processing on placental tissue. These comparisons confirm that the TriCleanse™ Process successfully removes the cellular component of the placenta while leaving the structural and modulating proteins intact in the final product. This is the goal of efficient decellularization.
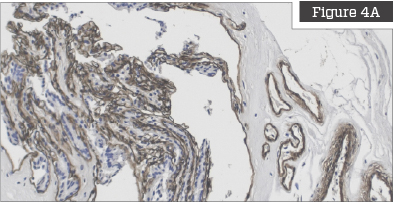
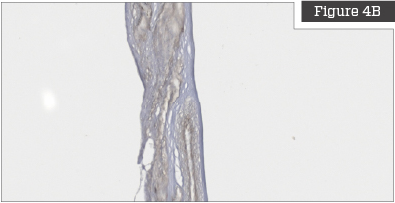
Cell and Cellular Debris Removal
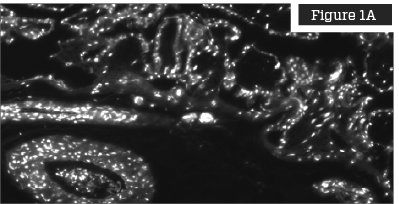
Figures on the left are untreated placentas and clearly show that the placenta is highly cellularized prior to processing. Figures on the right show placentas that have been treated with the TriCleanse™ Process; the treated placenta appears completely decellularized and void of any intact nuclei.
DAPI (4′, 6-diamidino-2-phenylindole) Staining
DAPI staining is a fluorescent DNA stain with high affinity for the AT region of double stranded DNA in either fixed or live cells.4 DAPI has been demonstrated to efficiently label cell nuclei for easier quantification.5
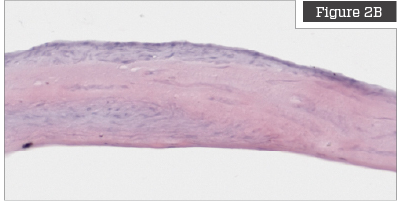
Hematoxylin & Eosin Staining
Hematoxylin and eosin (H&E) staining is commonly used to stain tissue for histology evaluation.6 H&E stains nuclei blue and ECM and cytoplasm pink and other tissues shades in between.
Preservation of the ECM
These images demonstrate that Collagen I, Collagen III, and laminin are present in the placenta prior to processing and remain in the tissue after processing. This is an indication that the TriCleanse™ Process has preserved the native structural proteins of the ECM.
Figures on the left show untreated placentas, and figures on the right show placentas treated with the TriCleanse™ Process.
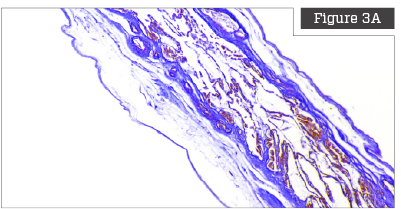
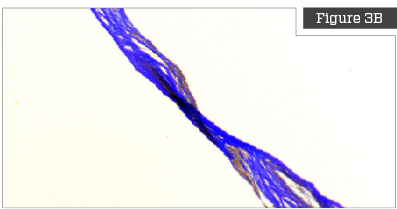
Herovici Staining
Herovici staining is a technique that utilizes two separate preferential chromatic stains and image processing to calculate the ratio of Collagen Type III to Collagen Type I.
Laminin Immunostaining
Immunostaining is a technique that allows for detection of specific proteins in the tissue. The staining allows for determination of presence and location of the protein. This particular stain was for laminin, a protein that is the major component of basement membranes.
Maintaining High Standards
Triad’s production facility was specifically designed to meet the rigorous ISO 13485 standards, including ISO Class 7 clean rooms, and the facility is staffed with highly trained technicians. This ensures that each product not only meets regulatory requirements, but also maintains Triad’s highest standards of safety and quality.
References
1. Gilpin, A., & Yang, Y. (2017). Decellularization Strategies for Regenerative Medicine: From Processing Techniques to Applications. Biomed Res Int, 2017, 9831534. doi:10.1155/2017/9831534
2. Keane, T. J., Londono, R., Turner, N. J., & Badylak, S. F. (2012). Consequences of ineffective decellularization of biologic scaffolds on the host response. Biomaterials, 33(6), 1771-1781. doi:10.1016/j.biomaterials.2011.10.054
3. Sicari, B. M., Dziki, J. L., Siu, B. F., Medberry, C. J., Dearth, C. L., & Badylak, S. F. (2014). The promotion of a constructive macrophage phenotype by solubilized extracellular matrix. Biomaterials, 35(30), 8605-8612. doi:10.1016/j.biomaterials.2014.06.060
4. Chazotte, B. (2011). Labeling nuclear DNA using DAPI. Cold Spring Harb Protoc, 2011(1), pdb prot5556. doi:10.1101/pdb.prot5556
5. Lau, Y. S., Xu, L., Gao, Y., & Han, R. (2018). Automated muscle histopathology analysis using CellProfiler. Skelet Muscle, 8(1), 32. doi:10.1186/s13395-018-0178-6
6. Hinton, J. P., Dvorak, K., Roberts, E., French, W. J., Grubbs, J. C., Cress, A. E., . . . Nagle, R. B. (2019). A Method to Reuse Archived H&E Stained Histology Slides for a Multiplex Protein Biomarker Analysis. Methods Protoc, 2(4). doi:10.3390/mps2040086